Market Spotlight: Single Cell Analysis, the Convergence of Microfluidics and “Omics” Platforms
The study of cells has been instrumental in understanding biological processes. Research on genes and proteins expressed in specific types of cells has typically been performed on samples containing many cells, with analysis reflecting the data averaged across them. But over the last several years, studies have indicated differences in cells of the same type or within the same population. Consequently, researchers are conducting single-cell analysis to study the unique qualities of individual cells.
Although individual cells have been analyzed microscopically for quite some time to study cell morphology and behavior, more recent interest in genomics and proteomics has generated demand for analysis of genes and proteins within single cells. Development of tools for single-cell genomic studies has progressed, with the ability to amplify both DNA and RNA of individual cells, facilitating targeted DNA sequencing, miRNA analysis and gene-expression studies. Advances in flow cytometry, which uses fluorescent probes to detect expression of specific proteins in cells, have extended the capacities of the technique to better resolve expression of gene products in single cells. Such advances in technology have enabled progression of single-cell analysis that provides unprecedented capabilities for systematically investigating cellular heterogeneity.
The intense interest of life science vendors in these developments is the motivation behind our latest report, The 2017 Market for Single Cell Analysis Products: Convergence of Microfluidics and Omics Platforms. One of the first things we learned was that researchers define “single cell analysis” differently. When we asked our scientists if they perform , single cell analysis those who answered yes were asked, “What BEST describes the nature of the single cell analysis work you perform?” This is how 1,157 scientists answered:
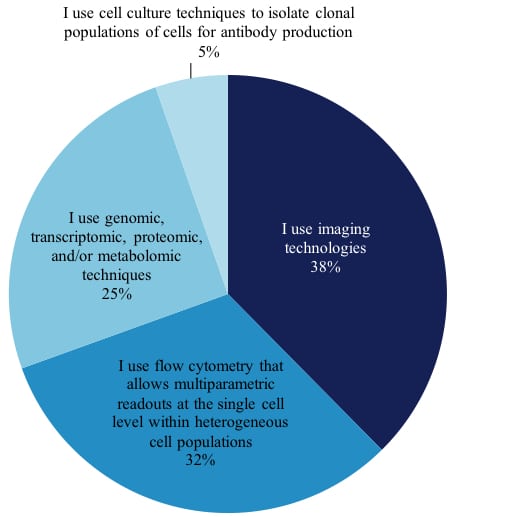
Techniques of Current Single Cell Analysis Researchers (excerpt from report)
We also asked 765 scientists whom plan to adopt single cell analysis in the next 12-24 months which techniques they plan to use and the distribution of techniques remains very similar to those in use today.
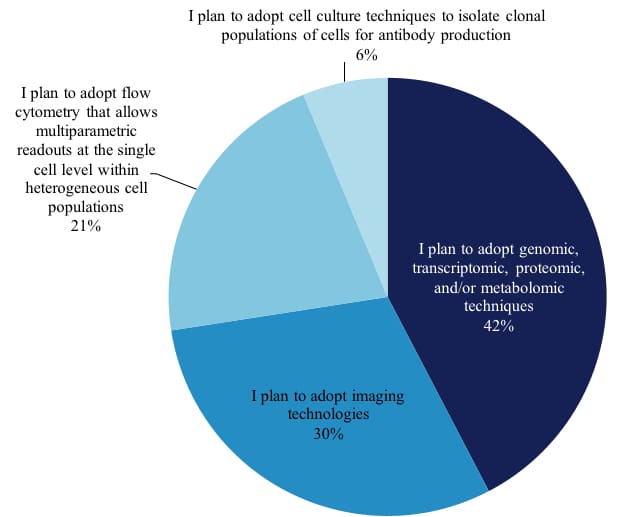
Techniques of Researchers Planning To Conduct Single Cell Analysis Researchers (excerpt from report)
For the purpose of our research, single cell analysis encompasses the application of genomic, transcriptomic, proteomic, and/or metabolomic techniques to the analysis of isolated, individually partitioned single cells. These techniques, coupled with new methods for cell isolation/partitioning, support the interrogation of single cells at increased levels of scale and throughput than previously possible.
Before a single cell can be analyzed in must be reliably isolated. To isolate a single cell from a heterogeneous population that preserves biological integrity, many scientists continue to use tried-and-true methods including micromanipulation, laser-capture microdissection and fluorescence-activated cell sorting (FACS); there are increasing number of new products on the market to isolate single cells with greater speed and throughput.
Isolation of single cells using microfluidics coupled with transcriptome analysis or mass cytometry emphasize analysis of many single cells in a single run. Several vendors offer technology for this, including the microfluidics-based C1™ Single-Cell Analysis System from Fluidigm, the DEPArray™ NxT System from Menarini Silicon Biosystems, and the Chromium™ Single Cell Controller from 10x Genomics.
One of our respondents sees isolation as a significant bottleneck:
Relating the molecular analysis of an isolated cell to its tissue location [physiology]/function is critical. While downstream molecular analyses are becoming easier by the day, the very beginning, namely the isolation of cells that the researcher can confidently know where they are coming from remains the biggest bottleneck in single cell investigations.
Unfortunately sources of error can creep into the workflow during amplification of the genetic material, at least a few cycles of amplification are usually necessary when studying the tiny amounts of RNA contained in a single cell. This could happen, for example, in RNA amplification when studying single-RNA transcriptomes; the different RNA levels that result could mistakenly be interpreted as cell-to-cell differences. RNA-Seq methods that employ molecular barcodes are designed to eliminate or minimize that type of experimental error. Furthermore, false positives can pop up during DNA amplification, because DNA polymerases are inherently error-prone. One scientist told us it is critically important to have good knowledge of the error rates associated with a particular method prior to investigating and reporting on biological variation. Many scientists in the field hope that their suppliers will deliver improvements in single-molecule sequencing will enable us to measure mutations in single cells directly without amplification.
Scientists have found that the average RNA expression of a group of cells is not necessarily the same as the RNA expression of a single cell from the same population. For instance, some genes are expressed continuously at a low level, while others are expressed strongly for only a brief period of time. Such patterns of expression become averaged out in macroscopic measurements—and are a significant driver of single cell analysis—the desire to reveal and understand, at the level of a single cell, the diversity of expression that is masked in the analysis of many cells simultaneously.
An advance in RNA sequencing known as RNA-capture-seq uses tiling arrays to “zoom in” on part of the transcriptome prior to sequencing. This gives better resolution and focuses the sequencing only on the area of interest. Agilent Technologies, Illumina, Roche Nimblegen and others offer target-enrichment tools for RNA sequencing. These target enrichment tools have been instrumental in overcoming barriers for work in single-cell analysis and are especially well-suited for clinical-research applications such as circulating tumor cells or single cells biopsied from embryos. Translational researchers are also in heterogeneous tumor samples, where it can be very important to look at one cell at a time.
Scientists are pushing suppliers for new approaches to single cell analyses that they need to better understand fundamental biological principles. The research community is looking for new analytical measures and manipulations of cellular contents, structure, and activity at the single cell level significantly beyond what is currently available. We received dozens of comments to an open-ended question asking our respondents what they considered the biggest barrier to their single cell research. Their responses are presented in the full report, but here are a few to consider:
3′ bias and depth of sequencing. A lot of genomic regions of interest can be identified by single cell sequencing, but establishing that these elements are “real” differences and that this event is a statistically significant difference for cells isolated for a given perturbation is not really easy to do and leads to a lot of false positives for regions of interest.
Another scientist commented:
Analyzing single cells using omic techniques is challenging because loss of material causes dropouts in the sequence and because sequencing errors are difficult to distinguish from real mutations. Any “omic” technique relies of the fact that it should be unbiased and large enough to give a true global picture of what is happening at the cellular. We are really limited by the throughput to be able to sample enough cells of a tumor to demonstrate that we have been able to genomically reconstruct the entire heterogeneity of the tumor such that we can now explain and predict the evolution of the tumor based on the genomics of the cellular population.
Cost was a concern of many researchers:
Dissociation of heterogeneous tissues giving a decent yield of viable cells. If you want to routinely do studies that involve hundreds of thousands of cells, even at an end-to-end cost of $0.10 per cell, that translates to $10,000 to $100,000 per experiment. The cost ultimately needs to be $0.01 per cell for use of the technology to become routine by most research labs.
According to the NIH, research based on single cell analysis will benefit from the following solutions:
- Devices and reagents to perform novel total (i.e., “-omic”) molecular and/or functional analyses of a wide variety of cell types (e.g., imaging-based spatial “-omics”, microdroplet-based high throughput “-omics”).
- Combinations of tools for multiplex analysis and/or manipulation of single cells to maximize data content over many parameters (e.g., gene expression, protein interaction, metabolic states, electrochemical dynamics, signal secretion/reception/transduction, cell adhesive properties, intercellular communication, cytoarchitectonic or migratory changes).
- Tools that provide significant advances in assay sensitivity, selectivity, scalability, or spatiotemporal resolution of molecules/structures/activities of single cells in situ.
- Automated manipulation or precise perturbation for scalable analysis of single cells, including parallel readouts in multiple cells and/or speed of processing.
- Tools and technologies that enable and transform single cell analysis in clinical tissue biopsies.
- Systems-level single cell dataset analysis or modeling, including computational approaches, in the context of tissues or whole organisms.
These needed technologies and tools must be developed in the context of a variety of experimental scenarios, such as:
- Identification of spatiotemporal transitions in cellular states (e.g., progenitor lineage determination, cellular aging, clonal selection and evolution, asymmetric division, cell specification, cell reprogramming).
- Detection or isolation of rare cells in a population (e.g., stem cells, tumor-initiating or metastatic cells, drug resistant cells).
- Elucidation of the cell molecular signatures at levels including DNA, RNA, protein, or metabolite) or functional consequences of molecular changes (e.g., genomic stability, epigenetic regulation, RNA modification or editing, protein modification or interaction, lipid metabolism)
- Characterization of heterogeneous cell responses to environmental changes (e.g., homeostatic perturbation, modulation of niche/microenvironment, morphogens or cell-to-cell signaling, toxic exposure, experience-dependent plasticity, host cell responses to infectious, immunological or allergic challenges).
The 2017 Market for Single Cell Analysis Products: Convergence of Microfluidics and Omics Platforms brings a much needed end-user perspective to the development of new products to support this fast growing segment of the market.